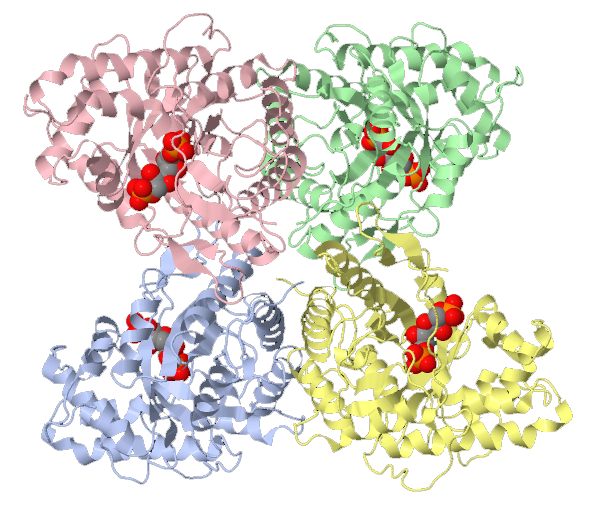
Lactate dehydrogenase reduces pyruvate to lactate and in the process oxidizes NADH back to NAD+ conditions. In Eukaryotes, lactate dehydrogenase is a tetrameric enzyme. Animals have two separate genes that code for two separate polypeptides that generate either the muscle (M) or heart (H) isoforms.
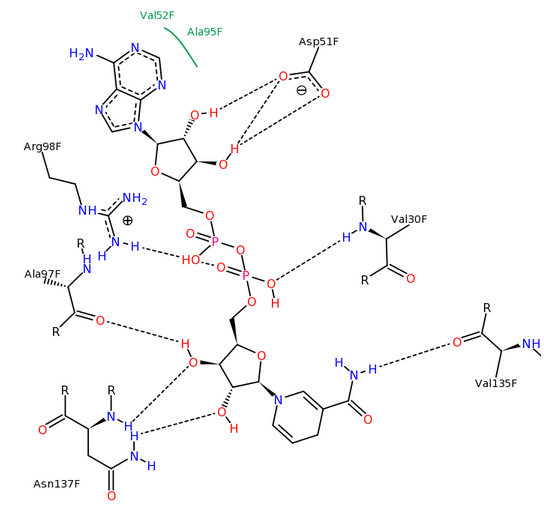
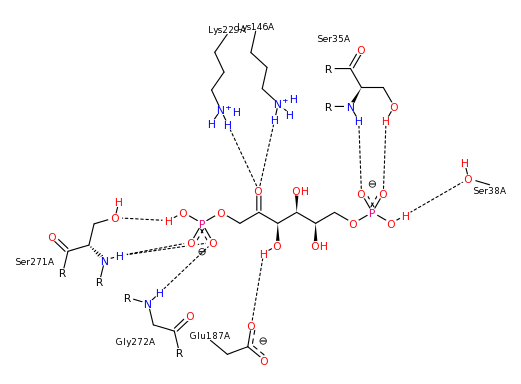
The binding of NADH by lactate dehydrogenase, generated with PoseView.
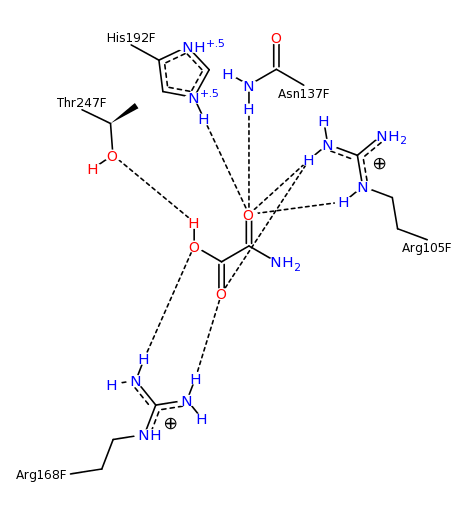
The binding of the inhibitor oxamic acid by lactate dehydrogenase. Image generated using PoseView.
Different combinations of M and H isoforms (e.g. H3M1) lead to different isozymes and different activities in different tissues. This difference in activity is due to a difference in Km for pyruvate which is dictated by the active site histidine (His 192). The pKa of this histidine differs by as much as 0.94 pH units between the muscle and heart isoforms, and results in the different kinetic s observed
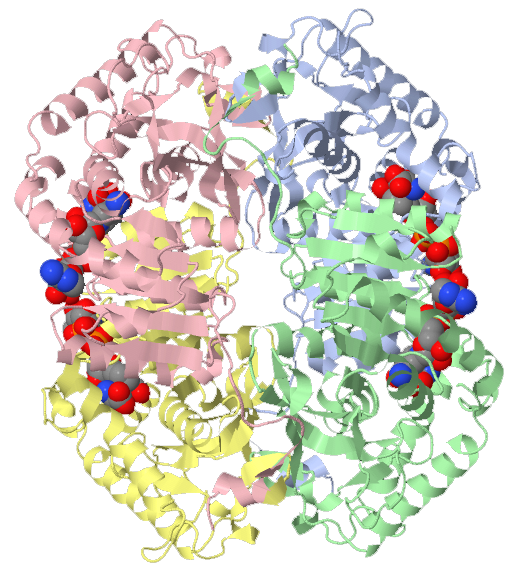
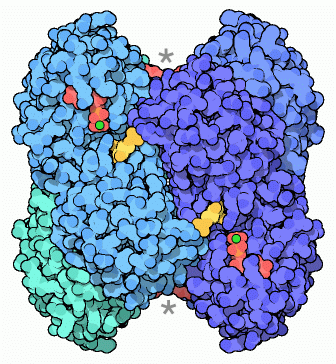
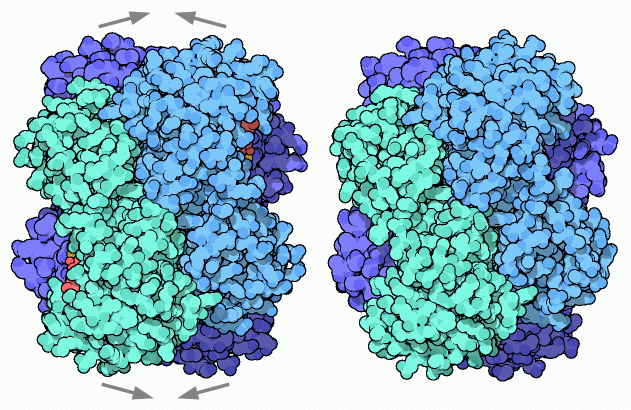
Biological assembly of human heart LDH bound to NADH and the inhibitor oxamic acid.
Lactate dehydrogenase is a mixed α/β protein and consists of a mixture of helix and sheet. Each subunit of lactate dehydrogenase folds into two discrete domains. The amino terminus forms what is known as a Rossman fold, a conserved domain found in many proteins that bind to NAD+ or NADP+. The carboxy terminus folds into a domain known as a lactate dehydrogenase fold, a domain conserved among all lactate dehydrogenases that binds to pyruvate.
Over 140 structures of lactate dehydrogenase have been determined. Many of these structures are bound to inhibitors of the enzyme. Plasmodium falciparum, the parasite that causes malaria uses lactate dehydrogenase to survive in the erythrocyte. This enzyme has been identified as a target for possible drug treatments for malaria, hence the interest in the structure of the complexes.